A University of Nebraska–Lincoln researcher is using National Science Foundation funding to build a first-of-its-kind tool that will help scientists better understand RNA, a molecule historically marginalized as DNA’s little sister but increasingly recognized as a vital biological force with great potential to impact human health and advance technology.
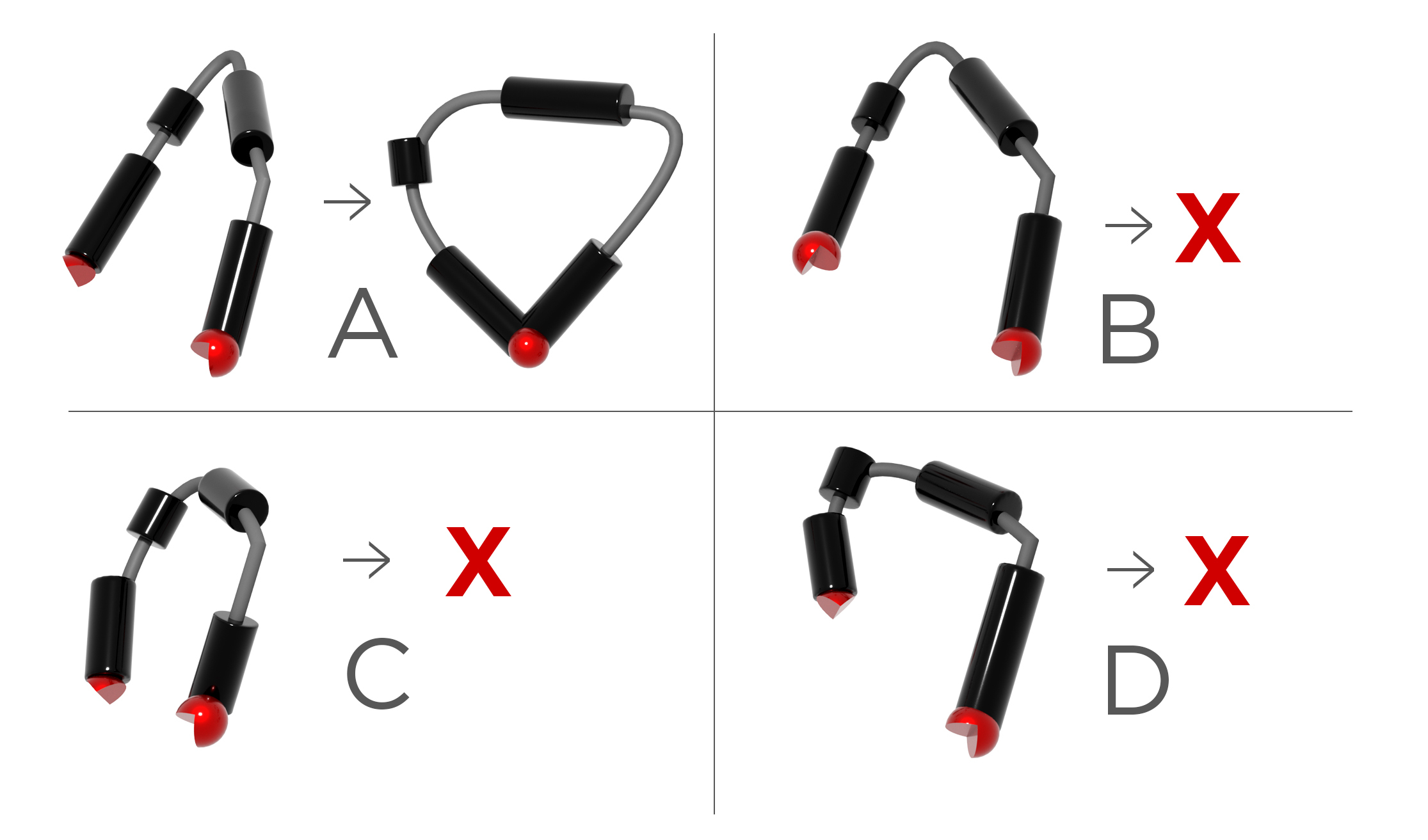
Joseph Yesselman, a Husker chemist, recently received a five-year, $1.2 million grant from the NSF Faculty Early Career Development Program, which he will use to develop the first computational model that can reliably predict RNA’s capacity to form tertiary contacts. These contacts, which are formed between RNA structural components known as motifs, enable RNA to fold into the complex 3D structures that determine their biological relevance.
The ability to quickly and easily predict whether an RNA forms tertiary contacts will help scientists shed light on the fundamental RNAs located inside cells and guide efforts to design new, stable RNAs useful in biotechnology, research and medicine.
The tool would also boost scientists’ ability to understand and manipulate RNA to improve human health. Many of the viruses most vexing to humans — influenza, COVID-19, Zika and rhinoviruses, to name a few — are RNA viruses, meaning their genetic material is made of RNA. Gleaning details about their 3D structure might provide hints about how to interrupt their survival and replication.
“Specifically for viruses, if you could disrupt one of the tertiary contacts, that might serve as a really interesting drug,” said Yesselman, assistant professor of chemistry.
Tertiary connections can form only when two conditions are fulfilled. First, an RNA strand must have specific types of motifs, which are discrete components that play a key role in RNA’s biological function and folding capacity. All RNAs have motifs, but just a tiny fraction of them are capable of making tertiary contacts — and there are no rules to determine which those are.
Second, even if the RNA has viable motifs, the strand must be malleable enough to bring them close together in physical space so they can link. It’s not always possible: Sometimes, the RNA segment is too short or too inflexible for the motifs to unite. Currently, there is no method for determining when a connection is possible.

“It’s really two separate ideas: Which motifs can come together and which strands are flexible enough to bring them together,” Yesselman said. “Models for both of these things don’t exist and are very difficult to navigate.”
To generate the data that will support his model, Yesselman is using an approach called mutational correlated chemical mapping. It’s a high-throughput technique where RNA sequences are tethered together, then disturbed through a series of mutations. By determining what mutation occurred, and how that mutation impacts other molecules in the structure, Yesselman will be able to pinpoint the motifs that are able to form tertiary contacts.
“You can see that a mutation caused reactivity somewhere else, and that those two points must be linked in space,” he said. “That’s enough to define what a tertiary contact is.”
To determine an RNA strand’s ability to bring the motifs together, he’ll capitalize on RNAMake, a system he previously developed to design and optimize 3D RNA structure. Yesselman will string together RNA segments, then analyze their reactions to mutations to determine whether the strand is flexible enough to unite partner motifs.
With this chemical mapping data, Yesselman will build an algorithm for the RNAMake software suite that computationally predicts the likelihood of tertiary contact formation. He’ll test the model by incorporating it into a new RNA interface for Foldit, an online puzzle video game focused on protein folding.
As part of his educational plan, he’ll launch other RNA 3D design challenges through the Foldit interface, allowing citizen scientists to contribute to the development of new RNA sequences. His laboratory will test the sequences, reporting the results back to the players.
Yesselman’s use of Foldit reflects his belief in harnessing the power of video games to engage students and the public in RNA science. This approach stems from his own life experiences. Yesselman said he recalls playing a video game as a young child that involved moving up a mountain, catching gnomes on the way up. To continue moving, you had to answer a science question. He’d go to the library, researching answers to the questions so he could advance. But it didn’t feel like learning in the conventional sense.
“That really stuck with me,” he said. “You can make things fun, and people can learn at the same time. You don’t need to know you’re doing science; it’s just a game. I think this whole movement of gamifying knowledge is really powerful.”
To bring the approach to Nebraska graduate and undergraduate students, Yesselman is developing a new course that uses video games and graphic user interfaces to introduce students to molecular design. He aims to make the daunting task of creating new RNA proteins a fun, accessible experience for students.
The National Science Foundation’s CAREER award supports pre-tenure faculty who exemplify the role of teacher-scholars through outstanding research, excellent education and the integration of education and research.
Share
Related Links
Tags
High Resolution Photos

HIGH RESOLUTION PHOTOS