University of Nebraska-Lincoln chemist Xiao Cheng Zeng recently conducted a simple scientific test during lunch at a local Chinese restaurant. He poured a few ounces of water onto the table top to see if the surface was water-loving (hydrophilic) or water-repellant (hydrophobic).
“This one I would say pretty much loves the water,” said Zeng, the Chancellor’s University Professor of Chemistry, as the water spread to cover a section several inches in diameter.
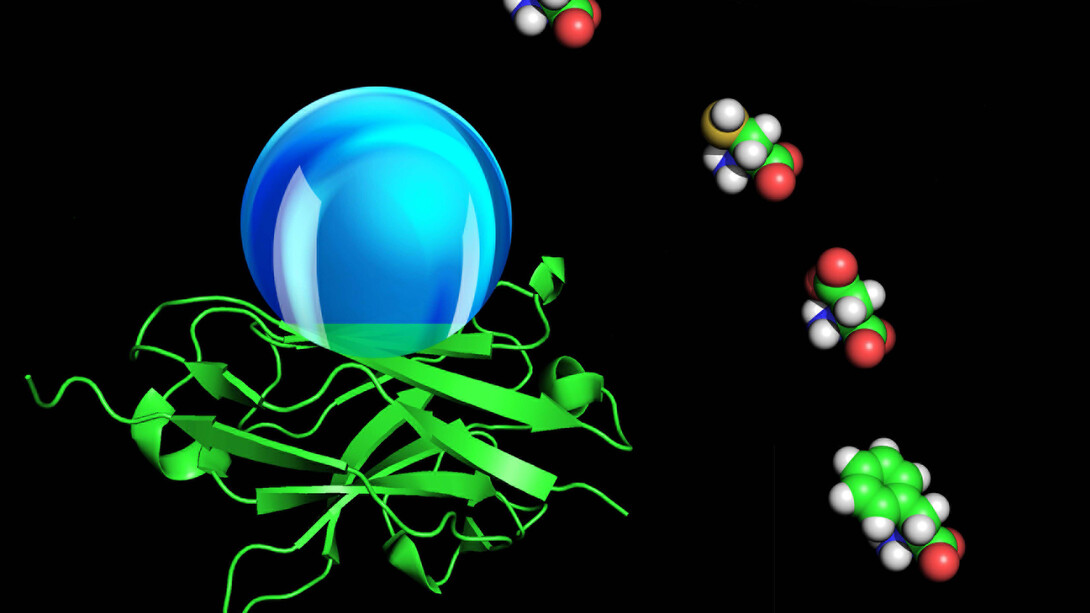
Far more complicated, however, are the findings of his recent paper on the hydrophobicity of proteins. Zeng and his co-authors recently published their computational data on the molecular hydrophobicity of amino acid chains in the Proceedings of the National Academy of Sciences.
Hydrophobic interactions help drive protein folding, a necessary step for the molecules to perform their biological functions.
“Misfolded proteins could cause diseases such as mad cow disease,” said Zeng, whose research is supported by the National Science Foundation.
The work of Zeng and his associates could help researchers decipher the role of hydrophobic interactions in protein folding, but measurements are critical. In the macro-environment of a kitchen countertop or a frying pan, engineers can optically measure the contact angle of a water droplet. The contact angle is the angular distance from the top of the surface to the top of the droplet.
“If the angle is big, then that means water doesn’t like to wet the surface,” Zeng explained. “If the angle is zero degrees, the water pretty much wets the entire surface.”
But the optical approach that engineers use to measure the contact angles on large, flat, wet surfaces doesn’t work well for proteins, which are microscopic and three-dimensional. Protein chemists measure hydrophobicity instead by studying the complex interactions between proteins and water. Zeng, however, sought to devise a measurement of protein hydrophobicity that he could share with engineers.
To do this, Zeng and colleague Joseph Francisco, the Elmer H. and Ruby M. Cordes Chair in Chemistry and dean of the College of Arts and Sciences, collaborated with Chinese physicists to create a network of proteins on a flat, artificial surface in a computer simulation.
“We stretch them and put them together side by side and then align them into a network,” Zeng explained. In the final stage, they add a virtual droplet of water to the mix to see what happens.
A single protein could contain millions of amino acid groups. In the past, however, chemists could measure only one such group per protein. But the Zeng-Francisco team’s work at Nebraska’s Holland Computing Center knows no such limitation.
“We have applied this computational measurement to all 20 types of amino acids in nature,” Zeng said, resulting in a new amino acid hydrophobicity scale.
Going for gold
Zeng’s theoretical and computational research spans energy science, materials science and atmospheric chemistry. A second paper published online Dec. 2 in Nature Communications displays his research versatility. In that paper, Zeng and three co-authors propose a grand unified model for understanding the structural richness of the 71 liganded gold clusters now known.
Ligands are ions or molecules that permit the clusters to form different species in solution. A better understanding of the atomic structure of the clusters could accelerate the development of potential applications.
“Liganded gold clusters have attracted intensive interest over the past 10 years owing to their broad and practical implications in catalysis, electrochemistry, quantum electronics and biomedicine,” Zeng and his co-authors wrote. The team performed supporting molecular computation at supercomputing centers in China and at the Nebraska Cluster for Computational Chemistry.
Nobel laureate Roger Kornberg of Stanford University first determined the structure of a big liganded gold cluster using X-ray crystallography in 2007. His work ignited new interest in the field.
Zeng’s latest paper represents a milestone in liganded gold cluster research, Francisco said.
“This is a major contribution,” said Francisco, one of Zeng’s frequent collaborators.
Zeng and his associates borrowed an idea from the world of high-energy physics for their classification scheme. In high-energy physics, quarks, the smallest known constituent of matter, come in six types or flavors: up, down, strange, charm, top and bottom. Now the field of liganded gold clusters has three flavors of its own: bottom, middle and top. These flavors represent valence states, which categorize the clusters’ ability to chemically unite with one another.
When gold clusters are small, they are like children who exhibit a different look and behavior as they age from 2 or 3 years old to 4 or 5, Zeng said. Similarly, a liganded gold cluster consisting of four atoms has a different structure and properties than a cluster consisting of five.
This structure-property relationship is important only for clusters ranging in size from two atoms up to about 1,000. At that point, the clusters are like children who have matured into adulthood and behave like regular gold, Zeng said.
Of the 71 known liganded gold clusters, scientists have crystallized the structures of 54 in the laboratory. Researchers have computationally predicted the remaining 17.
“We have 54 but maybe there are 100 or 200 to go,” Zeng said. Zeng predicts that his team’s new grand unified model will be able to accommodate them all.