To understand the disparity between the fields of education research and genomics, just consider how each might define the word “coding.”
To Nebraska’s Marilyne Stains, whose research on STEM education recently earned her the Presidential Early Career Award for Scientists and Engineers, it means classifying the classroom behavior of instructors and students.
To Robert Erdmann, who earned his doctorate studying plant genetics before joining Stains’ lab, it describes how organisms store the biological instruction manuals that make life possible.
But Stains brought Erdmann aboard precisely because his academic background differed from her own, prizing the chance to add a distinct perspective and voice to her lab. That investment in the interdisciplinary has paid off in the form of Classroom as Genome, a genetics-inspired approach that the duo developed to better analyze and interpret the data collected from classrooms.
The statistical and visualization tools that have accelerated the search for genetic indicators of disease or fitness in organisms have the potential to do the same for STEM education, the researchers said.
“I think the big innovation here is (being) able to leverage tools that are already vetted and existing in completely different fields and apply them to education data,” Stains, an associate professor of chemistry, said. “The tools that we are using here help us identify patterns of instructor (and) student behavior that we really could not do with just traditional statistics.”

In comparing notes while brainstorming the approach, Stains and Erdmann identified a few critical but easily overlooked similarities between genomes and classrooms.
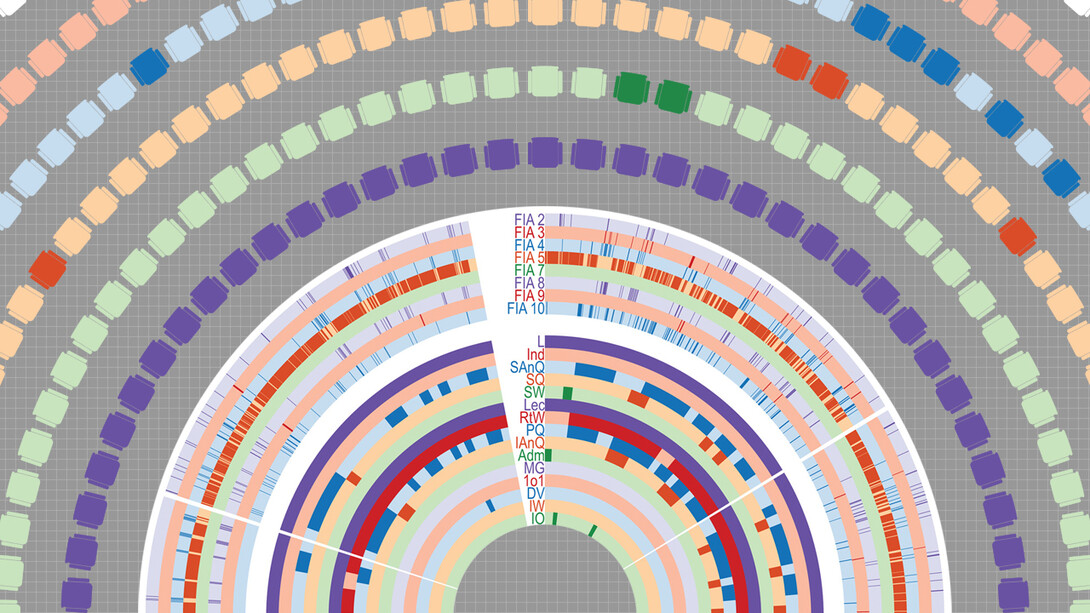
The duo realized, for instance, that both feature multiple layers of information which can get lost or compressed when looking only at the whole. Collectively, a genome can be thought of as the entire catalog of genetic blueprints in an organism. Understanding the genome on a practical level, though, means digging into the deeper layers: what DNA and genes actually are, how the instructions embedded in genes get transcribed and translated, why that process sometimes fails.
Most traditional approaches to analyzing classroom data resemble the former more than the latter, the researchers said, missing dynamics that sometimes best capture how instructors teach and students learn. Stains and Erdmann wanted the nuances. They wanted an approach that could account for both the influence of sequence — how one element might cause or affect the next — and the interplay among events that occur simultaneously or overlap in time. And they wanted to discern meaningful patterns from massive amounts of data gathered from hundreds or even thousands of classrooms.
Geneticists have faced similar yet even greater challenges while studying the genomes of organisms, many of which contain millions if not billions of nucleotide bases — the four “letters” of DNA’s alphabet. The technology-driven rise of bioinformatics over the past few decades has allowed geneticists to interpret the equivalents of words, pages and chapters in the instruction manuals formed by that code — along with the syntax, punctuation and other rules that determine how it gets transcribed.
In Erdmann’s mind, that staggering progress also represented unrealized potential.
“What I was seeing was an opportunity to use those same bioinformatics tools that I used for plant biology for a kind of unique and creative purpose: to analyze data that actually has a lot of parallels with biological data but hadn’t been viewed in that context before,” said Erdmann, now at the University of Minnesota Rochester. “I think we were both very pleased with how seamlessly it ended up being in terms of using those and the results we were able to get as we tested the tools.”
Ahead of the class
A major advantage of the Classroom as Genome approach, Stains and Erdmann said, is that it can incorporate multiple ways of measuring the same classroom observations. One common instrument, known as COPUS, helps categorize the presence or absence of behaviors and interactions in the classroom. Other instruments classify the perceived quality or other aspects of those events.
Education researchers typically analyze data from different instruments independently of one another, Stains said. But the new approach will allow researchers to layer the presence, quantity and quality of a practice or interaction into one visualization tool, giving them a more comprehensive but still comprehensible view of an instructor’s style or classroom culture, she said.
“Classrooms are chaotic places,” Erdmann said. “You want to be able to get as much information as you can and not lose any of it. This is a great data structure to use for that.
“This lets researchers use the best parts of multiple tools simultaneously to get more information out of the same set of data.”
To illustrate the use and value of Classroom as Genome, Stains and Erdmann included examples and case studies — the latter with data from a 2015 paper — when unveiling their approach in the journal CBE - Life Sciences Education.
Their examples consisted of questions that education researchers might better address using the approach, alongside the genomic equivalents already being answered via bioinformatics. A question on how evenly clicker questions are dispersed across a class period was equated with how far a piece of genetic code resides from other instances of the same code in a genome.
In a related case study, the duo used COPUS data and a genomics visualization tool to test the hypothesis that instructors issuing clicker questions also encourage students to collaborate before answering. Stains and Erdmann then expanded the analysis to demonstrate the breadth of related questions or hypotheses the approach could address.
“I think that will be particularly useful for education researchers or others who don’t know anything about these techniques,” Stains said. “If you use bioinformatics already, the language and the ways of thinking are probably common. But especially for folks who are outside that world, it was really important to demonstrate what these tools look like (and) what they can do.
“It’s kind of a proof-of-concept to see the potential that these methods have. But I think this is so new that we have to exemplify it.”
Stains and Erdmann said they hope the inverse will also prove true — that bench scientists who are more familiar with analyses in the lab than the classroom will come to appreciate and potentially use the latter.
“This could be a great bridge between folks in academic departments who are on the more biological end and folks who are thinking more along the educational lines,” Erdmann said. “If you provide opportunities for their worlds to collide, it can be very useful for both sides in terms of thinking about education in new ways and helping to make education research something that more people are thinking about.”
The researchers received support from the National Science Foundation.







