Tomas Helikar maneuvers his cursor to one among dozens of buttons neatly arranged in columns on his screen. He clicks it.
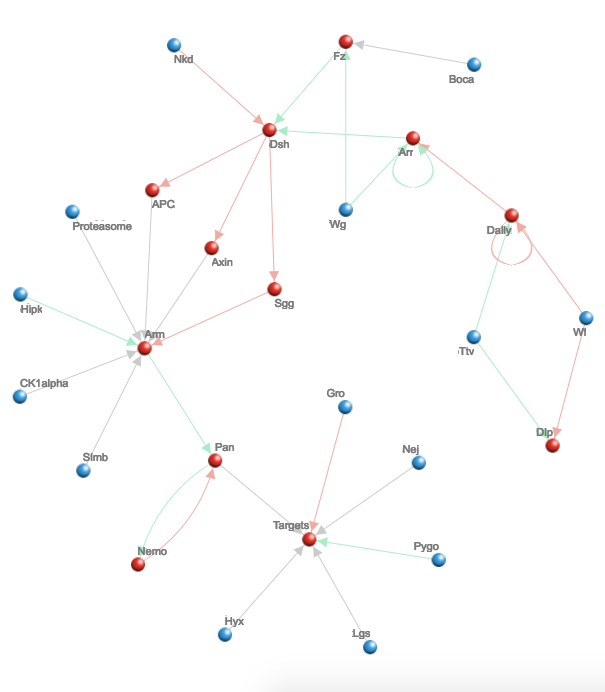
Almost immediately, several nodes in the vast graphical network that occupies the rest of Helikar’s monitor begin to shift from green to purple to red. Those nodes signify cellular proteins, he explains, and the digital redshift he has unleashed – a shift now spreading to still more nodes, these farther from the original – represents the cascading deactivation of those proteins by HIV.
Helikar, a computational biochemist at the University of Nebraska-Lincoln, runs a research lab dedicated to developing tools that mathematically capture the dynamics of both healthy and diseased biological systems. He also wants to make those tools as accessible as possible, useful and available even to those with no background in computer science or advanced mathematics.
For the second consecutive year, Helikar’s lab is advancing that goal by participating in the Google Summer of Code. The initiative, which began in 2005, provides paid opportunities for students around the globe to develop open-source software by collaborating with academic institutions and other organizations.
This summer, the Helikar Lab will work with five college students – three from India, one from Helikar’s native Czech Republic, one enrolled at Harvard – on a total of four projects designed to help organize, visualize and analyze the oceans of data that pour forth from biological processes.
“It helps our group to have an impact beyond UNL and beyond our local community,” said Helikar, assistant professor of biochemistry. “I came from a computer science and computer engineering background before I switched to a bioinformatics major. For me, it was very eye-opening to be able to see the potential impact that my computer science background could make in life sciences research. Being able to provide a similar opportunity for students is something that I get excited about.”
Helikar will directly oversee three of his lab’s Summer of Code projects. One aims to expand the functionality of his network visualization tool, a Web-based interface that can feature upward of 10,000 nodes when depicting especially complex biochemical processes.
“Visualizing large biological networks – or any networks – and interacting with them in a Web browser can be difficult because of the technologies available,” Helikar said. “There are some biology-specific tools that already allow you to do it, but their limit is the size of the network that you can visualize and smoothly interact with. In our research, we’re interested in studying as large of a system as possible.”
The interface already offers interactivity: Users can quickly move around a network and zoom in or out on the fly. But two students will craft Javascript components that allow users to edit text; select, add or delete individual nodes and connectors; and represent multiple values in a single node by altering textures and colors.
Another project will piggyback on a technology developed by Jiri Adamec, associate professor of biochemistry. Known as a Noviplex card, the technology can separate plasma from a finger-pricked blood sample that is blotted on the card. It also contains data on how concentrations of certain molecules in the blood affect the color intensity of plasma.
The associated coding project should yield a smartphone app that can visually read out analyses of the card’s plasma. By relying on a smartphone’s camera and processors, the application will compare a sample’s color against a baseline to determine how much the plasma’s molecular makeup deviates from normal concentrations. That information, in turn, could help prescreen for maladies ranging from cancer to the Zika virus.
And postdoctoral researcher Akram Mohammed will oversee the expansion of a Web-based platform that makes statistical analyses less daunting to students and researchers who lack the command-line knowledge of statistical programming languages such as R.
“We want students and researchers who don’t know R, or are just getting into understanding how it works, to be able to start with the actual analysis,” Helikar said. “People seem to have more fun playing with something that already functions first, and then figuring out how it works, than the other way around.
“Students in life sciences have varying degrees of knowing programming and being intimidated by programming. We’re interested in being able to provide … a scaffolding where you can upload your data and do your analysis – just point and click. But there will also be a window where you can say, ‘Show me the R code that generated this graph.’ The next step would be changing some parameters in that code to see how it affects the graph. And the final step would be, ‘OK, now make more substantial changes to the R code or write your own to see what kind of analyses you can do.’”
This same spirit of accessibility previously spurred Helikar to create Cell Collective, a Web-based software library that allows researchers to construct large-scale models of biological systems and run real-time simulations to study their dynamics.
Whether modeling gene regulation within an organism or food webs across an ecosystem, the platform lets researchers run hundreds of experimental simulations in a matter of minutes. Though the complex mathematical equations underpinning those models are readily available, users can also run the simulations without ever manually entering or modifying the equations.
“In biomedical research and life sciences research, computational modeling has been used for a very long time,” Helikar said. “But I realized that (it) has been utilized mostly by computational scientists – those who can write code and mathematical equations. My dream is that laboratory researchers will be able to … generate new hypotheses and revise their existing hypotheses by building and simulating these models, just the way they use pipettes in the lab on a day-to-day basis.
“You don’t want to make it a complete black box that nobody can dive into if they want, but you don’t need to be a mechanic to be able to drive your car. You need to understand that if you press the gas pedal, it’s going to go, and if you press the brake pedal, it’s going to stop. You need to have these assumptions and understand those principles, but you don’t need to understand how the engine works. That’s how I look at computational modeling. I want to bring that to the hands of laboratory researchers.”